Editorial: %p2PSA and PHI improve diagnostic accuracy for prostate cancer
Serum samples from patients with prostate cancer (PCa) are measured routinely for total PSA (tPSA) and complexed PSA as a part of the diagnostic evaluation. In addition, a variety of molecular isoforms of the PSA molecule can be assayed, including free PSA (fPSA), %freePSA, [-2]proPSA, %[-2]proPSA and the new combined calculation, termed the Prostate Health Index (PHI), which is calculated using [-2]proPSA, fPSA and tPSA. The PHI was developed by Beckman Coulter immunoassays and is performed on Access-2- or DxI800-instruments. Numerous clinical studies have been conducted with the isoforms of PSA (fPSA, %fPSA, [-2]proPSA and %[-2]ProPSA) to test the PCa diagnosis and prognosis predictions, and to predict the need for rebiopsies [1]. In addition to these various molecular isoforms of US Food and Drug Administration-approved clinical bioassays for PSA, several PSA-computed derivatives have also been developed to assess kinetics such as PSA velocity, PSA density and PSA doubling time, and all of these tests have been used to evaluate mostly prognostic events (i.e. disease progression, metastasis and death) in patients with PCa [2].
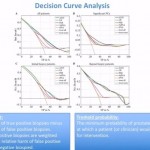
In the recent publication by Boegemann et al. in BJUI [3], the PHI is calculated as ([-2]proPSA/fPSA) × √tPSA). The PHI has been investigated previously with regard to its use in the diagnosis and prognosis of PCa, and key findings have included its role in defining PCa tumour aggressiveness [1, 4, 5], its value in predicting active surveillance upstaging [6, 7], and its ability to predict the need for a rebiopsy [1]. When authors decide that a new PSA test may be valuable in the management of PCa, they usually include historic PSA and PSA isoforms for statistically relevant comparisons. It is most gratifying, therefore, that Boegemann et al. studied a total of 769 men ≤ 65 years old, scheduled for initial or repeat prostate biopsy and that they were recruited from four test sites. Their in-depth analysis included total PSA as well as its isoforms, generating univariate analysis. Results for %[-2]proPSA and the PHI yielded similar areas under the curve (0.72 and 0.73, respectively) in all patients (P < 0.001). They also showed that the PHI had the best performance in predicting the initial biopsy (0.67 and 0.68, respectively) and repeat biopsy (0.79 and 0.78, respectively) results. To substantiate their observations, the authors tested several multivariate models using logistic regression and artificial neural network (ANN) methods. Both worked, and in multivariate analysis PHI was added to a base model that included age, prostate volume and DRE results, tPSA and %fPSA. The PHI was strongest in predicting PCa in all patients, at initial and repeat biopsy and for significant PCa (AUC 0.73, 0.68, 0.78 and 0.72, respectively). All results were strongly supported by decision-curve analysis tools for ANN and logistic regression modelling, as well as single variable analysis for the PHI, %fPSA, tPSA, ‘treat all’ and ‘treat none’ data.
In summary, the multicentre study by Boegemann et al. assessed 769 men aged ≤ 65 years at their initial and repeat prostate biopsy diagnoses and %[-2]proPSA and PHI were shown to be the strongest predictors of biopsy outcome. The two multivariate prediction ANN models (BM-1 and BM-2) were shown to minimize the risk of missing clinically significant PCa, while reducing the number of unnecessary biopsies in men without PCa or potentially insignificant disease.
Currently, evaluation of high-risk cancer is often based on genomic knowledge and has ~70–75% accuracy to offer personalized treatment regimens. The present manuscript achieved similar accuracy using the PHI and routine clinicopathological features to create models for PCa detection and repeat biopsy decision-making. Furthermore, many patients and their doctors choose definitive treatment that might be unnecessary and can cause incontinence and/or impotence. Active surveillance is an alternative option for very-low- to low-risk PCa cases using a minimal number of positive biopsy cores, PSA density ≥and tPSA ≥ 10 ng/mL for entry criteria [6, 7]. Clearly, a PCa risk predictor containing the best biomarkers, including the PHI, will improve the accuracy in the management of patients on active surveillance. There is also a need at the pretreatment diagnostic step to determine whether the pathological stage (i.e. non-organ-confined status) of the tumour requires definitive treatment (i.e. surgery and/or irradiation), with or without adjuvant therapy. Hence, we will need the best clinicopathological quantitative medical imaging and histomorphological (molecular and histological) information at pretreatment stage to create new and more accurate nomograms or tables and have them validated. Ultimately, the patient requires risk models with an accuracy close to 100% to make treatment decisions safely and with confidence [8].