Posts
Article of the week: A transcriptomic signature of tertiary Gleason 5 predicts worse clinicopathological outcome
Every week, the Editor-in-Chief selects an Article of the Week from the current issue of BJUI. The abstract is reproduced below and you can click on the button to read the full article, which is freely available to all readers for at least 30 days from the time of this post.
In addition to the article itself, there is an editorial written by a prominent member of the urological community, and a video produced by the authors. These are intended to provoke comment and discussion and we invite you to use the comment tools at the bottom of each post to join the conversation.
If you only have time to read one article this week, it should be this one.
A transcriptomic signature of tertiary Gleason 5 predicts worse clinicopathological outcome
Abstract
Objective
To investigate the genomic features of tertiary pattern 5 (TP5) on radical prostatectomy specimens in an effort to explain the poor clinical outcomes associated with this disease subtype.
Patients and methods
Data from 159 men with Gleason Grade Group (GGG) 3 or 4 were considered. All patients had Decipher diagnostic testing with transcript profiles and single‐channel array normalisation (SCAN)‐normalised expression of coding genes.
The relationship between Decipher and TP5 was investigated by linear and binary logistic regressions. A differential transcriptomic analysis between patients with and without TP5 was performed. The prognostic role of these genes on progression‐free survival (PFS) and overall survival (OS) was evaluated using The Cancer Genome Atlas.
Results
In all, 52/159 (33%) patients had GGG 3–4 with TP5 disease. TP5 was associated with a higher Decipher score (β 0.07, 95% confidence interval [CI] 0.02–0.13; P = 0.04) and higher likelihood of falling within the intermediate‐ or high‐risk categories (odds ratio 3.34, 95% CI 1.34–8.35; P = 0.01). Analysis of microarray data revealed an 18‐gene signature that was differentially expressed in patients with TP5; 13 genes were over‐ and five under‐expressed in the TP5 cohort.
The overexpression of cyclin dependent kinase inhibitor 2B (CDKN2B), polo‐like kinase 1 (PLK1), or cell division cycle 20 (CDC20) was associated with worse PFS. The group harbouring overexpression of at least one gene had a 5‐year PFS rate of 50% vs 74% in the group without overexpression (P < 0.001).
Conclusions
Our studies have elucidated unique genomic features of TP5, whilst confirming previous clinical findings that patients harbouring TP5 tend to have worse prognosis. This is the first RNA‐based study to investigate the molecular diversity of TP5 and the first correlating CDKN2B to poorer prognosis in patients with prostate cancer.
Editorial: Transcriptomic signatures for tertiary pattern 5 in prostate cancer
Significant evidence is now emerging and publications have shown that the molecular characterization of tumours represents an important approach to stratifying patients with prostate cancer into appropriate treatments and their responses, helping to inform the next steps in the care pathway of these patients [1]. Understanding the functional role of the genes included in these signatures also gives an insight into the biology of the disease and how it develops and progresses.
BJUI has published papers in the past describing gene signatures that are associated with pathological Gleason score as pathological markers of aggressiveness and their potential role in predicting outcome [2].
The paper by Martini et al. [3] in the current issue of BJUI adds to this important literature. In a cohort of 159 patients, of whom 52 had tertiary pattern 5 (TP5) prostate cancer, Martini et al. showed that TP5 was associated with higher risk categories in the Decipher genomic score. Their study supports the literature showing that patients with TP5 have an elevated likelihood of developing disease after radical prostatectomy and have a poor prognosis [4]. It should be noted, however, that this is a small cohort with few patients defined as having TP5 disease. As prostate cancer is highly heterogeneous and gene expression can also be affected by the patient’s background, additional validation of this finding will be required in independent cohorts.
Additional analysis of the 698 gene by Martini et al. [3] identified a unique RNA signature of 18 genes for the TP5 cohort. A limitation of using the 698 gene previously demonstrated to be related to prostate cancer is that those with TP5 disease may be a unique subset of patients that may be associated with unfamiliar pathological pathways and new genes. An examination of novel genes whose expression pattern is unique to patients with TP5 disease may reveal additional genes.
Martini et al. then used the independent TCGA provisional database to link the expression of these 18 genes with clinical features. Patients harbouring any of the three genes CDKN2B, PLK1 and CDC20 mRNA were found to have worse progression‐free survival. The authors go on to undertake analysis of the 18 genes using the ingenuity network analysis tool identifying pathways involved in the cell cycle, DNA replication and repair, cellular assembly, cell death and survival and gene expression, which would fit with the biology of progressive disease. They also revealed a role in the dedifferentiation process and progression of cells towards anaplasia. They specifically identified CDKN2B, which has previously been associated with tumour suppression but might actually be linked to tumour progression, and identified it for further investigation.
Finally the researchers found that 13 genes out of the 18‐gene TP5 signature are upregulated in tumour lesions with TP5. Following these results they performed a sensitivity analysis in which the expression values of the genes in the cohort with GGG3‐4 and TP5 were compared with those of patients with GGG5 and discovered that these genes were similarly overexpressed in both groups. This finding strengthens the hypothesis that these genes are implicated in the dedifferentiation process of tumour cells, but it also raises the question of whether the pattern of expression of these genes is unique to patients with TP5 only, or whether it is associated with every tumour (primary, secondary or tertiary) that is classified as GGG5. Further research is needed to understand what the clinical significance of this genetic signature is and what pathological process it describes in practice.
This investigation into the functional role of the genes has given an insight into the biology of prostate cancer progression and TP5 disease which might inform a future area of research for novel biomarker panels and therapeutic interventions.
by R. William Watson and Omri Taltsh
References
- , , , , , , Genomic markers in prostate cancer decision making. Euro Urol 2018; 73: 572– 82
- , , , , , , , ,, , , , Evaluation of a 24‐Gene signature for prognosis of metastatic events and prostate cancer‐specific mortality. BJU Int 2017; 119: 961– 7
- , , , , , , ,, , A transcriptomic signature of tertiary Gleason 5 predicts worse clinicopathological outcome. BJU Int 2019; 124: 155– 62
- , , , , , Significance of tertiary Gleason pattern 5 in Gleason score 7 radical prostatectomy specimens. J Urol 2008; 179:516– 22
Video: A transcriptomic signature of tertiary Gleason 5 predicts worse clinicopathological outcome
A transcriptomic signature of tertiary Gleason 5 predicts worse clinicopathological outcome
by Alberto Martini
Abstract
Objective
To investigate the genomic features of tertiary pattern 5 (TP5) on radical prostatectomy specimens in an effort to explain the poor clinical outcomes associated with this disease subtype.
Patients and methods
Data from 159 men with Gleason Grade Group (GGG) 3 or 4 were considered. All patients had Decipher diagnostic testing with transcript profiles and single‐channel array normalisation (SCAN)‐normalised expression of coding genes.
The relationship between Decipher and TP5 was investigated by linear and binary logistic regressions. A differential transcriptomic analysis between patients with and without TP5 was performed. The prognostic role of these genes on progression‐free survival (PFS) and overall survival (OS) was evaluated using The Cancer Genome Atlas.
Results
In all, 52/159 (33%) patients had GGG 3–4 with TP5 disease. TP5 was associated with a higher Decipher score (β 0.07, 95% confidence interval [CI] 0.02–0.13; P = 0.04) and higher likelihood of falling within the intermediate‐ or high‐risk categories (odds ratio 3.34, 95% CI 1.34–8.35; P = 0.01). Analysis of microarray data revealed an 18‐gene signature that was differentially expressed in patients with TP5; 13 genes were over‐ and five under‐expressed in the TP5 cohort.
The overexpression of cyclin dependent kinase inhibitor 2B (CDKN2B), polo‐like kinase 1 (PLK1), or cell division cycle 20 (CDC20) was associated with worse PFS. The group harbouring overexpression of at least one gene had a 5‐year PFS rate of 50% vs 74% in the group without overexpression (P < 0.001).
Conclusions
Our studies have elucidated unique genomic features of TP5, whilst confirming previous clinical findings that patients harbouring TP5 tend to have worse prognosis. This is the first RNA‐based study to investigate the molecular diversity of TP5 and the first correlating CDKN2B to poorer prognosis in patients with prostate cancer.
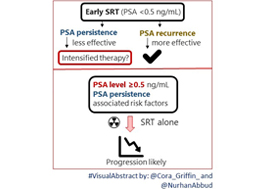
Article of the week: Nomogram helps the preoperative prediction of early biochemical recurrence after radical prostatectomy
Every week the Editor-in-Chief selects the Article of the Week from the current issue of BJUI. The abstract is reproduced below and you can click on the button to read the full article, which is freely available to all readers for at least 30 days from the time of this post.
In addition to the article itself, there is an accompanying editorial written by a prominent member of the urological community. This blog is intended to provoke comment and discussion and we invite you to use the comment tools at the bottom of each post to join the conversation.
Finally, the third post under the Article of the Week heading on the homepage will consist of additional material or media. This week we feature a video of Ángel Borque discussing his paper.
If you only have time to read one article this week, it should be this one.
Genetic predisposition to early recurrence in clinically localized prostate cancer
Ángel Borque, Jokin del Amo†, Luis M. Esteban*, Elisabet Ars§, Carlos Hernández**, Jacques Planas¶, Antonio Arruza††, Roberto Llarena‡, Joan Palou§, Felipe Herranz**, Carles X. Raventós¶, Diego Tejedor†, Marta Artieda†, Laureano Simon†, Antonio Martínez†, Elena Carceller, Miguel Suárez, Marta Allué¶, Gerardo Sanz* and Juan Morote¶
‘Miguel Servet’ University Hospital, *University of Zaragoza, Zaragoza, Spain, †Progenika Biopharma S.A., ‡University Hospital of Cruces, Bilbao, §Puigvert Foundation, ¶‘Vall d’Hebron’ University Hospital, Barcelona, **‘Gregorio Marañón’ University Hospital, Madrid, and ††Hospital of Txagorritxu, Vitoria, Spain
OBJECTIVES
• To evaluate genetic susceptibility to early biochemical recurrence (EBCR) after radical prostatectomy (RP), as a prognostic factor for early systemic dissemination.
• To build a preoperative nomogram to predict EBCR combining genetic and clinicopathological factors.
PATIENTS AND METHODS
• We evaluated 670 patients from six University Hospitals who underwent RP for clinically localized prostate cancer (PCa), and were followed-up for at least 5 years or until biochemical recurrence.
• EBCR was defined as a level prostate-specific antigen >0.4 ng/mL within 1 year of RP; preoperative variables studied were: age, prostate-specific antigen, clinical stage, biopsy Gleason score, and the genotype of 83 PCa-related single nucleotide polymorphisms (SNPs).
• Univariate allele association tests and multivariate logistic regression were used to generate predictive models for EBCR, with clinicopathological factors and adding SNPs.
• We internally validated the models by bootstrapping and compared their accuracy using the area under the curve (AUC), net reclassification improvement, integrated discrimination improvement, calibration plots and Vickers’ decision curves.
RESULTS
• Four common SNPs at KLK3, KLK2, SULT1A1 and BGLAP genes were independently associated with EBCR.
• A significant increase in AUC was observed when SNPs were added to the model: AUC (95% confidence interval) 0.728 (0.674–0.784) vs 0.763 (0.708–0.817).
• Net reclassification improvement showed a significant increase in probability for events of 60.7% and a decrease for non-events of 63.5%.
• Integrated discrimination improvement and decision curves confirmed the superiority of the new model.
CONCLUSIONS
• Four SNPs associated with EBCR significantly improved the accuracy of clinicopathological factors.
• We present a nomogram for preoperative prediction of EBCR after RP.
Read Previous Articles of the Week
Editorial: Prostate cancer families – predicting disease before and after the radical
In this issue of BJUI, Borque et al. discuss a subject that is now very close to my heart. Aged 48 years, I am 6 weeks post radical prostatectomy for a Gleason 3 + 4 prostate adenocarcinoma measuring ~2 mL in volume, with a PSA level of 2.54 ng/mL. Histology reassures me it is organ confined and seminal vesicle negative. My father and his brother both died aged 63 years of Gleason 10 prostate cancer and my brother is awaiting his radical prostatectomy in a few weeks. I have two sons, one of whom has asked me when he should be tested. Any prognostic information is going to help me advise my family.
In all, 85% of prostate cancers appear to be sporadic. The incidence of all prostate cancers is 1 in 8500 under the age of 40 years, rising to 1 in 15 at 60–69 years and 1 in 8 after that. The lifetime risk in the UK for all men is 8–10%.
The genetics of prostate cancer are confused by case clustering; the family members of men with a prostate cancer diagnosis seek out early advice from their physician resulting in detection of some clinically questionable cancers and an apparent higher incidence in certain families. These families do not necessarily have genetically determined prostate cancer.
The lifetime risk is altered dramatically by having two or more first-degree relatives with a diagnosis of prostate cancer; if the disease in the relative is identified before the age of 65 years the risk is increased further. Bratt suggests the risk rises from 15 to 20% when a single first-degree relative is diagnosed aged < 60 years. Zeegers et al., in a meta-analysis, have shown that diagnosing prostate cancer in a relative aged < 65 years increases the relative risk of having prostate cancer by 3.3, and having two first-degree relatives increases the relative risk by a factor of 5.1.
Analysis of a huge database from Sweden including data on 182 000 fathers and 3700 sons with prostate cancer suggest a standardised incidence ratio of 9.4 in men with a father and brother diagnosed with prostate cancer, with further analysis also showing unsurprisingly that the risk increases as an individual ages. Some true ‘prostate cancer families’ have been identified. These families have three or more relatives with prostate cancer often associated with a diagnosis at a young age, possibly with an increased tendency to an aggressive
phenotype; my uncle was 18 months from diagnosis to death from his disease, my father 4 years. In these families, the relative risk in male family members is 3.39 in those where the diagnosis of identified sufferers was made aged > 65 years, and 7.33 where the diagnosis is in men aged < 65 years. These risks which effectively give a lifetime risk in the individual of 45–50% are associated with carriage of a gene identified as increasing the prostate cancer risk. The best identified of these genes is the BRCA2 (breast cancer type 2 susceptibility protein) gene, which is associated with an increased risk of other cancers including breast, ovarian, gallbladder and pancreatic cancer, as well as malignant melanoma. This gene, carried in 1% of
Ashkenazi Jewish families, is associated with prostate cancer families in this population.
Now my prostate has been removed, I need to determine my chance of treatment failure. It would be interesting to know whether my genes and my single nucleotide polymorphisms (SNPs), which have almost certainly been responsible for me developing prostate cancer, can also predict my chance of developing early biochemical recurrence (EBCR) and the possibility of needing further treatment. In the Borque et al. article, I would appear on the first model (Fig. 1) to have a chance of ECBR of between 1 and 5%. This risk, according to this study, could increase to up to 30%, if I was to have four SNPs associated with prostate cancer (Fig. 2). Furthermore, we need to know whether identification of SNPs is any better than other possible predictors of EBCR and disease progression, such as the identification of lymphovascular invasion and tumour volume in the final specimen and the presence of extraprostatic extension, data not included in this study. Incidentally, I had no evidence of lymphovascular invasion.
The authors identify that this study needs repeating, particularly in a more ethnically diverse group (this study included Caucasian origin as an entry criterion), and we await longer term data to see how SNPs predict metastasis and prostate cancer-related death.
Jonathan M. Glass
Department of Urology, Guys & St Thomas’ Hospital Trust, London, UK
Video: Genetic predisposition to early recurrence in clinically localized prostate cancer
Genetic predisposition to early recurrence in clinically localized prostate cancer
Ángel Borque, Jokin del Amo†, Luis M. Esteban*, Elisabet Ars§, Carlos Hernández**, Jacques Planas¶, Antonio Arruza††, Roberto Llarena‡, Joan Palou§, Felipe Herranz**, Carles X. Raventós¶, Diego Tejedor†, Marta Artieda†, Laureano Simon†, Antonio Martínez†, Elena Carceller, Miguel Suárez, Marta Allué¶, Gerardo Sanz* and Juan Morote¶
‘Miguel Servet’ University Hospital, *University of Zaragoza, Zaragoza, Spain, †Progenika Biopharma S.A., ‡University Hospital of Cruces, Bilbao, §Puigvert Foundation, ¶‘Vall d’Hebron’ University Hospital, Barcelona, **‘Gregorio Marañón’ University Hospital, Madrid, and ††Hospital of Txagorritxu, Vitoria, Spain
• To evaluate genetic susceptibility to early biochemical recurrence (EBCR) after radical prostatectomy (RP), as a prognostic factor for early systemic dissemination.
• To build a preoperative nomogram to predict EBCR combining genetic and clinicopathological factors.
PATIENTS AND METHODS
• We evaluated 670 patients from six University Hospitals who underwent RP for clinically localized prostate cancer (PCa), and were followed-up for at least 5 years or until biochemical recurrence.
• EBCR was defined as a level prostate-specific antigen >0.4 ng/mL within 1 year of RP; preoperative variables studied were: age, prostate-specific antigen, clinical stage, biopsy Gleason score, and the genotype of 83 PCa-related single nucleotide polymorphisms (SNPs).
• Univariate allele association tests and multivariate logistic regression were used to generate predictive models for EBCR, with clinicopathological factors and adding SNPs.
• We internally validated the models by bootstrapping and compared their accuracy using the area under the curve (AUC), net reclassification improvement, integrated discrimination improvement, calibration plots and Vickers’ decision curves.
RESULTS
• Four common SNPs at KLK3, KLK2, SULT1A1 and BGLAP genes were independently associated with EBCR.
• A significant increase in AUC was observed when SNPs were added to the model: AUC (95% confidence interval) 0.728 (0.674–0.784) vs 0.763 (0.708–0.817).
• Net reclassification improvement showed a significant increase in probability for events of 60.7% and a decrease for non-events of 63.5%.
• Integrated discrimination improvement and decision curves confirmed the superiority of the new model.
CONCLUSIONS
• Four SNPs associated with EBCR significantly improved the accuracy of clinicopathological factors.
• We present a nomogram for preoperative prediction of EBCR after RP.